Uncovering the Interplays between Cyanobacterial Blooms and Antibiotic Resistance Genes
Abstract
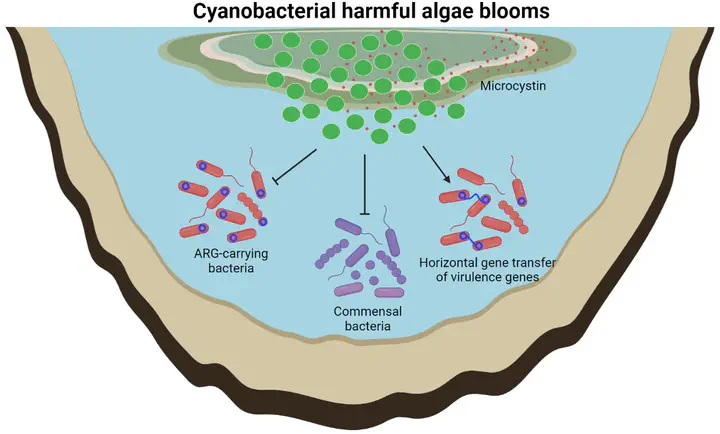
Cyanobacterial harmful algae blooms (cyanoHABs), which are a form of microbial dysbiosis of freshwater environments, are an emerging environmental and public health concern. In addition, the freshwater environment serves as a reservoir for antibiotic resistance genes (ARGs), which can pose a risk of transmission during microbial dysbiosis, such as cyanoHABs. However, the interactions between potential synergistic pollutants, cyanoHABs, and ARGs remain poorly understood. During cyanoHABs, a total of nine regions showed dominance of Microcystis and high microcystin levels. The resistome, mobilome, and microbiome were interrelated and lined to the physicochemical properties of freshwater. Planktothrix and Pseudanabaena exhibited competitive interactions with Actinobacteriota and Proteobacteria during cyanoHABs. A total of 42 ARG carriers were identified, most of which belonged to Actinobacteriota and Proteobacteria. ARG carriers showed a strong correlation with ARGs density, which decreased with the severity of cyanoHAB. Although ARGs decreased due to a reduction of ARG carriers during cyanoHABs, mobile gene elements (MGEs) and virulence factors (VFs) genes increased. We monitored the potential synergistic pollutions of cyanoHABs and ARGs propagation. Our findings demonstrated that cyanobacteria compete with freshwater commensal bacteria such as Actinobacteriota and Proteobacteria, which carry ARGs in freshwater, resulting in a reduction of ARGs. Moreover, cyanoHABs generate biotic and abiotic stress in the freshwater microbiome, which may leads to an increase in MGEs and VFs. The investigation of the intricate interplays between microbiome, resistome, mobilome, and pathobiome during cyanoHABs not only interprets the mechanisms underlying the dynamics of microbial dysbiosis but also emphasizes the imperative of prioritizing the prevention of microbial dysbiosis in the risk management of ARGs.